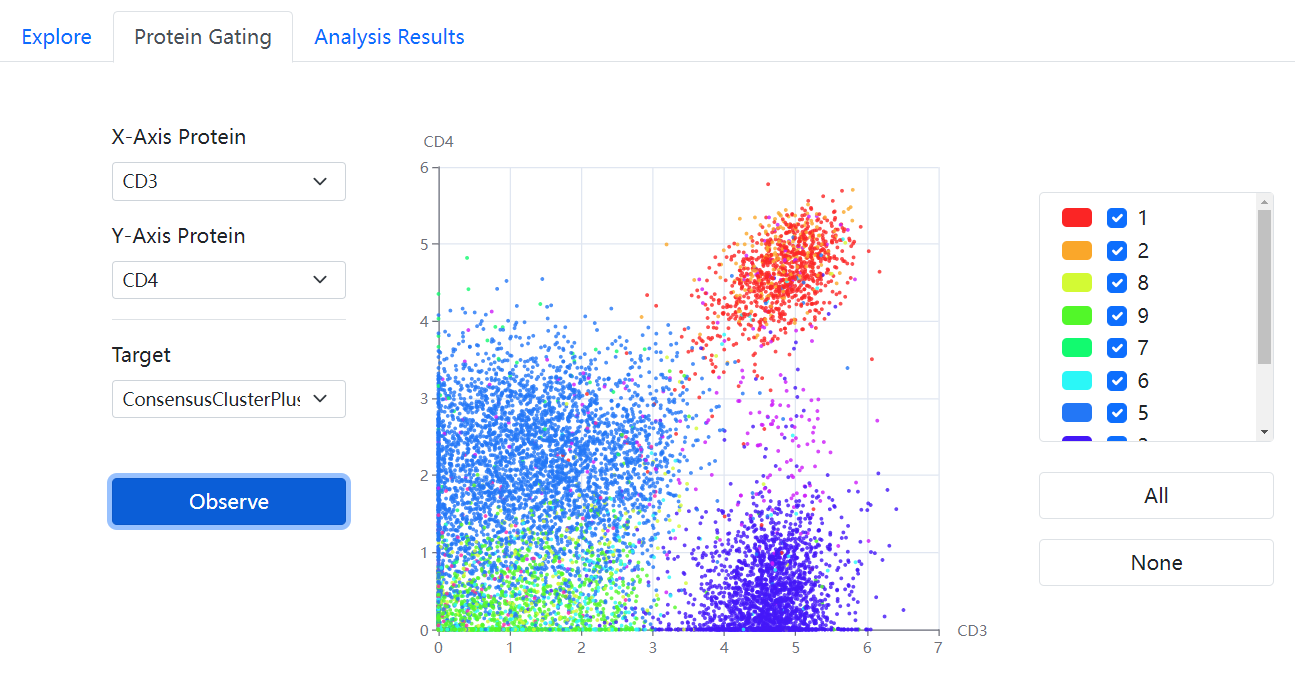
Single-cell Proteomic DataBase (SPDB) is a comprehensive repository
for general single-cell proteomic data, encompassing both antibody-based
and mass spectrometry-based single-cell proteomics, and provides
dataset/protein search, visualization and download with unified formats.